Data setup and configuration
Supported image formats
FAST uses bio-formats to open a range of different image formats. These include:
- Tagged image files (.tif/.tiff)
- Zeiss microscopy images (.czi)
- Zeiss laser-scanning microscopy images (.lsm)
- Nikon NIS-elements images (.nd2)
- Leica image file format images (.lif)
among many others. For a full list, see this page.
File structure
The FAST pipeline analyses each imaging dataset within a unique directory, referred to throughout this manual as the 'root' directory. Initially, this directory should be set up to contain a single image file, which can contain multiple channels and timepoints. Three-dimensional images consisting of multiple z-positions are not currently supported.
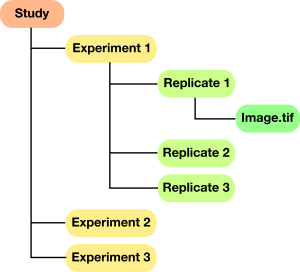
For large experiments consisting of many individual image series, each separate image file should be placed in a separate directory. For example, we may have collected data under several different experimental conditions, with several replicates of each condition. In this case we might set up our directory structure as shown:
Once a single image series has been analysed using the FAST GUI, other datasets within the experiment can be analysed using the same parameters with FAST's batch processing system.