coarsegrainTrackData.m
Class: Function
Description: Bins spatial coordinates into the specified grid and finds the instantaneous average velocity and orientation of all objects within these bins for each timepoint. This coarsegrained representation of the motility data is output.
Author: Oliver J. Meacock
- coarsegrainTrackData.m
function [fieldFlow,fieldPhi] = coarsegrainTrackData(procTracks,trackSettings,noBinsX,noBinsY) %COARSEGRAINTRACKDATA converts track data into continuum flowfield data by %binning up cells and averaging their instantaneous velocities in each bin. % % INPUTS: % -procTracks: The track data to be coarsegrained % -trackSettings: The settings used to generate the input track data % -noBinsX: The number of bins in the x-direction % -noBinsY: The number of bins in the y-direction % % OUTPUTS: % -fieldFlow: The coarse-grained flowfields % -fieldPhi: The coarse-grained orientation fields % % Author: Oliver J. Meacock, 2021 %Bin data from the PTV approach into boxes equivalent to the PIV grid xEdges = linspace(0,trackSettings.maxX,noBinsX+1); yEdges = linspace(0,trackSettings.maxY,noBinsY+1); flowBins = cell(noBinsX,noBinsY,trackSettings.maxF); oriBins = cell(noBinsX,noBinsY,trackSettings.maxF); %Collate data from all tracks into bin structure %Track loop for i = 1:size(procTracks,2) us = procTracks(i).vmag .* cosd(procTracks(i).theta); vs = -procTracks(i).vmag .* sind(procTracks(i).theta); %Negative undoes the flipping of theta in the original version of the data oris = procTracks(i).phi; ts = procTracks(i).times; for j = 1:size(procTracks(i).x,1)-1 xBin = find(diff(procTracks(i).x(j) > xEdges)); yBin = find(diff(procTracks(i).y(j) > yEdges)); tBin = ts(j); if ~isempty(xBin)&&~isempty(yBin) flowBins{yBin,xBin,tBin} = [flowBins{yBin,xBin,tBin};us(j),vs(j)]; oriBins{yBin,xBin,tBin} = [oriBins{yBin,xBin,tBin};oris(j)]; end end end fieldFlow = zeros(noBinsX,noBinsY,trackSettings.maxF,2); fieldPhi = zeros(noBinsX,noBinsY,trackSettings.maxF); for i = 1:noBinsX for j = 1:noBinsY for t = 1:fieldSettings.maxF fieldFlow(i,j,t,1) = nanmean(flowBins{i,j,t}(:,1)); fieldFlow(i,j,t,2) = nanmean(flowBins{i,j,t}(:,2)); fieldPhi(i,j,t) = rad2deg(circ_mean(deg2rad(oriBins{i,j,t})*2)/2); end end end
Example usage
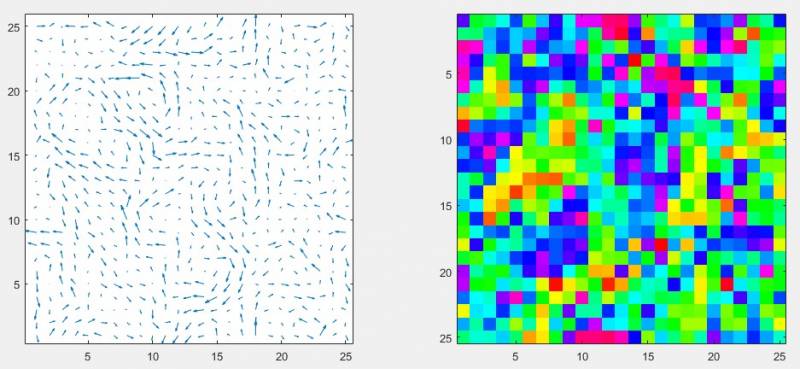
%Apply coarsegraining procedure to some input monolayer data [fieldFlow,fieldPhi] = coarsgrainTrackData(procTracks,fieldSettings,40,40) %Visualise results for 100th timepoint using quiver and image representations of flow and orientation data respectively subplot(1,2,1) quiver(fieldFlow(:,:,100,1),fieldFlow(:,:,100,2)) axis equal subplot(1,2,2) imagesc(fieldPhi(:,:,100)) axis equal colormap('hsv') %HSV is a circular colourmap - appropriate for use with circular data